GAparsimony package
GAparsimony module
Combines feature selection(FS), hyperparameter tuning (HT), and parsimonious model selection (PMS) with Genetic Algorithm (GA) optimization. GA selection procedure is based on separate cost and complexity evaluations. Therefore, the best individuals are initially sorted by an error fitness function, and afterwards, models with similar costs are rearranged according to the model complexity measurement so as to foster models of lesser complexity. The algorithm can be run sequentially or in parallel.
GAparsimonypackage is a new GA wrapper automatic method that efficiently generated machine learning models with reduced complexity and adequate generalization capacity.ga_parsimonyfunction is primarily based on combining FS and HT with a second novel GA selection process (named ReRank algorithm) in order to achieve better overall parsimonious models. Unlike other GA methodologies that use a penalty parameter for combining loss and complexity measures into a unique fitness function, the main contribution of this package is that ga_parsimony selects the best models by considering cost and complexity separately. For this purpose, the ReRank algorithm rearranges individuals by their complexity when there is not a significant difference between their costs. Thus, less complex models with similar accuracy are promoted. Furthermore, because the penalty parameter is unnecessary, there is no consequent uncertainty associated with assigning a correct value beforehand. As a result, with GAPARSIMONY, an automatic method for obtaining parsimonious models is finally made possible.
References
F.J. Martinez-de-Pison, J. Ferreiro, E. Fraile, A. Pernia-Espinoza, A comparative study of six model complexity metrics to search for parsimonious models with GAparsimony R Package, Neurocomputing, Volume 452, 2021, Pages 317-332, ISSN 0925-2312, https://doi.org/10.1016/j.neucom.2020.02.135.
Martinez-de-Pison, F.J., Gonzalez-Sendino, R., Aldama, A., Ferreiro-Cabello, J., Fraile-Garcia, E. Hybrid methodology based on Bayesian optimization and GA-PARSIMONY to search for parsimony models by combining hyperparameter optimization and feature selection (2019) Neurocomputing, 354, pp. 20-26. https://doi.org/10.1016/j.neucom.2018.05.136
Urraca R., Sodupe-Ortega E., Antonanzas E., Antonanzas-Torres F., Martinez-de-Pison, F.J. (2017). Evaluation of a novel GA-based methodology for model structure selection: The GA-PARSIMONY. Neurocomputing, Online July 2017. https://doi.org/10.1016/j.neucom.2016.08.154
Martinez-De-Pison, F.J., Gonzalez-Sendino, R., Ferreiro, J., Fraile, E., Pernia-Espinoza, A. GAparsimony: An R package for searching parsimonious models by combining hyperparameter optimization and feature selection (2018) Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 10870 LNAI, pp. 62-73. https://doi.org/10.1007/978-3-319-92639-1_6
Applications
Eduardo Dulce-Chamorro, Francisco Javier Martinez-de-Pison, An advanced methodology to enhance energy efficiency in a hospital cooling-water system, Journal of Building Engineering, Volume 43, 2021, 102839, ISSN 2352-7102,https://doi.org/10.1016/j.jobe.2021.102839.
Sanz-Garcia, A., Fernandez-Ceniceros, J., Antonanzas-Torres, F., Pernia-Espinoza, A.V., Martinez-De-Pison, F.J. GA-PARSIMONY: A GA-SVR approach with feature selection and parameter optimization to obtain parsimonious solutions for predicting temperature settings in a continuous annealing furnace (2015) Applied Soft Computing Journal, 35, art. no. 3006, pp. 13-28. https://doi.org/10.1016/j.asoc.2015.06.012
Fernandez-Ceniceros, J., Sanz-Garcia, A., Antoñanzas-Torres, F., Martinez-de-Pison, F.J. A numerical-informational approach for characterising the ductile behaviour of the T-stub component. Part 2: Parsimonious soft-computing-based metamodel (2015) Engineering Structures, 82, pp. 249-260. https://doi.org/10.1016/j.engstruct.2014.06.047
Antonanzas-Torres, F., Urraca, R., Antonanzas, J., Fernandez-Ceniceros, J., Martinez-De-Pison, F.J. Generation of daily global solar irradiation with support vector machines for regression (2015) Energy Conversion and Management, 96, pp. 277-286. https://doi.org/10.1016/j.enconman.2015.02.086
- class GAparsimony.gaparsimony.GAparsimony(fitness, params, features, type_ini_pop='improvedLHS', popSize=50, pcrossover=0.8, maxiter=40, feat_thres=0.9, rerank_error=0.0, iter_start_rerank=0, pmutation=0.1, feat_mut_thres=0.1, not_muted=3, tol=0.0001, elitism=None, selection='nlinear', keep_history=False, early_stop=None, maxFitness=inf, suggestions=None, seed_ini=None, verbose=1)
Bases:
object- __init__(fitness, params, features, type_ini_pop='improvedLHS', popSize=50, pcrossover=0.8, maxiter=40, feat_thres=0.9, rerank_error=0.0, iter_start_rerank=0, pmutation=0.1, feat_mut_thres=0.1, not_muted=3, tol=0.0001, elitism=None, selection='nlinear', keep_history=False, early_stop=None, maxFitness=inf, suggestions=None, seed_ini=None, verbose=1)
A class for searching parsimonious models by feature selection and parameter tuning with genetic algorithms.
- Parameters
fitness (function) – The fitness function, any function which takes as input a chromosome which combines the model parameters to tune and the features to be selected. Fitness function returns a numerical vector with three values:validation_cost, testing_cost and model_complexity, and the trained model.
params (dict) –
It is a dictionary with the model’s hyperparameters to be adjusted and the range of values to search for.
{ "<< hyperparameter name >>": { "range": [<< minimum value >>, << maximum value >>], "type": GAparsimony.FLOAT/GAparsimony.INTEGER }, "<< hyperparameter name >>": { "value": << constant value >>, "type": GAparsimony.CONSTANT } }
features (int or list of str) – The number of features/columns in the dataset or a list with their names.
type_ini_pop (str, {'randomLHS', 'geneticLHS', 'improvedLHS', 'maximinLHS', 'optimumLHS', 'random'}, optional) – Method to create the first population with GAparsimony._population function. Possible values: randomLHS, geneticLHS, improvedLHS, maximinLHS, optimumLHS, random.First 5 methods correspond with several latine hypercube for initial sampling. By default is set to improvedLHS.
popSize (int, optional) – The population size.
pcrossover (float, optional) – The probability of crossover between pairs of chromosomes. Typically this is alarge value and by default is set to 0.8.
maxiter (float, optional) – The maximum number of iterations to run before the GA process is halted.
feat_thres (float, optional) – Proportion of selected features in the initial population. It is recommended a high percentage of the selected features for the first generations. By default is set to 0.90.
rerank_error (float, optional) – When a value is provided, a second reranking process according to the model complexities is called by parsimony_rerank function. Its primary objective isto select individuals with high validation cost while maintaining the robustnessof a parsimonious model. This function switches the position of two models if the first one is more complex than the latter and no significant difference is found between their fitness values in terms of cost. Thus, if the absolute difference between the validation costs are lower than rerank_error they areconsidered similar. Default value=`0.01`.
iter_start_rerank (int, optional) – Iteration when ReRanking process is actived. Default=`0`. Sometimes is useful not to use ReRanking process in the first generations.
pmutation (float, optional) – The probability of mutation in a parent chromosome. Usually mutation occurswith a small probability. By default is set to 0.10.
feat_mut_thres (float, optional) – Probability of the muted features-chromosome to be one. Default value is set to 0.10.
not_muted (int, optional) – Number of the best elitists that are not muted in each generation. Default valueis set to 3.
elitism (int, optional) – The number of best individuals to survive at each generation. By default the top 20% individuals will survive at each iteration.
selection (str, optional) – Method to perform selection with GAparsimony._selection function. Possible values: linear, nlinear, random. By default is set to nlinear.
keep_history (bool, optional) – If it is True keeps in the list GAparsimony.history each generation as pandas.DataFrame. This parameter must set True in order to use GAparsimony.plot method or GAparsimony.importance function.
maxFitness (int, optional) – The upper bound on the fitness function after that the GA search is interrupted. Default value is set to +Inf.
early_stop (int, optional) – The number of consecutive generations without any improvement in the bestfitness value before the GA is stopped.
suggestions (numpy.array, optional) – A matrix of solutions strings to be included in the initial population.
seed_ini (int, optional) – An integer value containing the random number generator state.
verbose (int, optional) – The level of messages that we want it to show us. Possible values: 1=monitor level, 2=debug level, if 0 no messages. Default 1.
- population
The current (or final) population.
- Type
- minutes_total
Total elapsed time (in minutes).
- Type
float
- history
A list with the population of all iterations.
- Type
float
- best_score
The best validation score in the whole GA process.
- Type
float
- best_model
The best model in the whole GA process.
- best_model_conf
The parameters and features of the best model in the whole GA process.
- Type
- bestfitnessVal
The validation cost of the best solution at the last iteration.
- Type
float
- bestfitnessTst
The testing cost of the best solution at the last iteration.
- Type
float
- bestcomplexity
The model complexity of the best solution at the last iteration.
- Type
float
Examples
Usage example for a regression model using the sklearn boston dataset
from sklearn.model_selection import RepeatedKFold from sklearn.linear_model import Lasso from sklearn.preprocessing import StandardScaler from sklearn.metrics import mean_squared_error from sklearn.datasets import load_boston from GAparsimony import GAparsimony, Population, getFitness from GAparsimony.util import linearModels_complexity boston = load_boston() X, y = boston.data, boston.target X = StandardScaler().fit_transform(X) # ga_parsimony can be executed with a different set of 'rerank_error' values rerank_error = 0.01 params = {"alpha":{"range": (1., 25.9), "type": Population.FLOAT}, "tol":{"range": (0.0001,0.9999), "type": Population.FLOAT}} cv = RepeatedKFold(n_splits=10, n_repeats=10, random_state=42) fitness = getFitness(Lasso, mean_squared_error, linearModels_complexity, cv, minimize=True, test_size=0.2, random_state=42, n_jobs=-1) GAparsimony_model = GAparsimony(fitness=fitness, params = params, features = boston.feature_names, keep_history = True, rerank_error = rerank_error, popSize = 40, maxiter = 50, early_stop=10, feat_thres=0.90, # Perc selected features in first generation feat_mut_thres=0.10, # Prob of a feature to be one in mutation seed_ini = 1234) GAparsimony_model.fit(X, y) GAparsimony_model.summary() aux = GAparsimony_model.summary() GAparsimony_model.plot()
GA-PARSIMONY | iter = 0 MeanVal = -79.1813338 | ValBest = -30.3470614 | TstBest = -29.2466835 |ComplexBest = 13000000021.927263| Time(min) = 0.185549 GA-PARSIMONY | iter = 1 MeanVal = -55.0713465 | ValBest = -30.2283235 | TstBest = -29.2267507 |ComplexBest = 12000000022.088743| Time(min) = 0.1238126 GA-PARSIMONY | iter = 2 MeanVal = -34.8473723 | ValBest = -30.2283235 | TstBest = -29.2267507 |ComplexBest = 12000000022.088743| Time(min) = 0.0907046 GA-PARSIMONY | iter = 3 MeanVal = -38.5251529 | ValBest = -30.0455259 | TstBest = -29.2712578 |ComplexBest = 10000000022.752678| Time(min) = 0.0755356 ... GA-PARSIMONY | iter = 20 MeanVal = -34.2636095 | ValBest = -29.5036901 | TstBest = -29.3245069 |ComplexBest = 5000000023.115818| Time(min) = 0.0659549 GA-PARSIMONY | iter = 21 MeanVal = -40.4629864 | ValBest = -29.5036901 | TstBest = -29.3245069 |ComplexBest = 5000000023.115818| Time(min) = 0.0725066 GA-PARSIMONY | iter = 22 MeanVal = -35.9230384 | ValBest = -29.5036901 | TstBest = -29.3245069 |ComplexBest = 5000000023.115818| Time(min) = 0.0704362 GA-PARSIMONY | iter = 23 MeanVal = -36.5946762 | ValBest = -29.5036901 | TstBest = -29.3245069 |ComplexBest = 5000000023.115818| Time(min) = 0.0723252 GA-PARSIMONY | iter = 24 MeanVal = -37.3293511 | ValBest = -29.5036901 | TstBest = -29.3245069 |ComplexBest = 5000000023.115818| Time(min) = 0.0684883 +------------------------------------+ | GA-PARSIMONY | +------------------------------------+ GA-PARSIMONY settings: Number of Parameters = 2 Number of Features = 13 Population size = 40 Maximum of generations = 50 Number of early-stop gen. = 10 Elitism = 8 Crossover probability = 0.8 Mutation probability = 0.1 Max diff(error) to ReRank = 0.01 Perc. of 1s in first popu.= 0.9 Prob. to be 1 in mutation = 0.1 Search domain = alpha tol CRIM ZN INDUS CHAS NOX RM AGE DIS RAD \ Min_param 1.0 0.0001 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 Max_param 25.9 0.9999 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 1.0 TAX PTRATIO B LSTAT Min_param 0.0 0.0 0.0 0.0 Max_param 1.0 1.0 1.0 1.0 GA-PARSIMONY results: Iterations = 25 Best validation score = -29.502012171608403 Solution with the best validation score in the whole GA process = fitnessVal fitnessTst complexity alpha tol CRIM ZN INDUS CHAS NOX \ 0 -29.502 -29.3244 5e+09 1.33694 0.541197 0 0 0 0 0 RM AGE DIS RAD TAX PTRATIO B LSTAT 0 1 1 0 0 0 1 1 1 Results of the best individual at the last generation = Best indiv's validat.cost = -29.503690126221098 Best indiv's testing cost = -29.324506895493244 Best indiv's complexity = 5000000023.115818 Elapsed time in minutes = 2.031593410174052 BEST SOLUTION = fitnessVal fitnessTst complexity alpha tol CRIM ZN INDUS CHAS NOX \ 0 -29.5037 -29.3245 5e+09 1.3374 0.547189 0 0 0 0 0 RM AGE DIS RAD TAX PTRATIO B LSTAT 0 1 1 0 0 0 1 1 1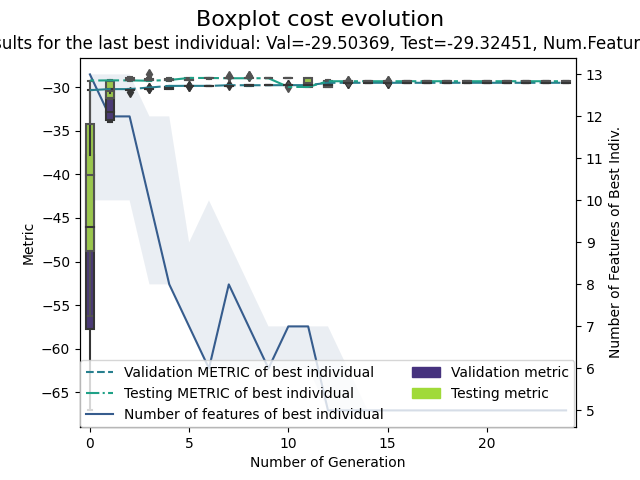
Regression plot
Usage example for a classification model using the wine dataset
from sklearn.model_selection import RepeatedKFold from sklearn.svm import SVC from sklearn.metrics import cohen_kappa_score from sklearn.datasets import load_wine from GAparsimony import GAparsimony, Population, getFitness from GAparsimony.util import svm_complexity wine = load_wine() X, y = wine.data, wine.target X = StandardScaler().fit_transform(X) rerank_error = 0.001 params = {"C":{"range": (00.0001, 99.9999), "type": Population.FLOAT}, "gamma":{"range": (0.00001,0.99999), "type": Population.FLOAT}, "kernel": {"value": "poly", "type": Population.CONSTANT}} cv = RepeatedKFold(n_splits=10, n_repeats=10, random_state=42) fitness = getFitness(SVC, cohen_kappa_score, svm_complexity, cv, minimize=False, test_size=0.2, random_state=42, n_jobs=-1) GAparsimony_model = GAparsimony(fitness=fitness, params=params, features=wine.feature_names, keep_history = True, rerank_error = rerank_error, popSize = 40, maxiter = 50, early_stop=10, feat_thres=0.90, # Perc selected features in first generation feat_mut_thres=0.10, # Prob of a feature to be one in mutation seed_ini = 1234) GAparsimony_model.fit(X, y) GAparsimony_model.summary() GAparsimony_model.plot()
GA-PARSIMONY | iter = 0 MeanVal = 0.879549 | ValBest = 0.9314718 | TstBest = 0.9574468 |ComplexBest = 10000000045.0| Time(min) = 0.1438692 GA-PARSIMONY | iter = 1 MeanVal = 0.9075035 | ValBest = 0.9496819 | TstBest = 0.9142857 |ComplexBest = 11000000060.0| Time(min) = 0.0893566 GA-PARSIMONY | iter = 2 MeanVal = 0.9183232 | ValBest = 0.9496819 | TstBest = 0.9142857 |ComplexBest = 11000000060.0| Time(min) = 0.0818844 GA-PARSIMONY | iter = 3 MeanVal = 0.9219764 | ValBest = 0.9534295 | TstBest = 0.9568345 |ComplexBest = 10000000043.0| Time(min) = 0.0739248 ... GA-PARSIMONY | iter = 19 MeanVal = 0.9182586 | ValBest = 0.972731 | TstBest = 1.0 |ComplexBest = 7000000048.0| Time(min) = 0.0867344 GA-PARSIMONY | iter = 20 MeanVal = 0.9224294 | ValBest = 0.972731 | TstBest = 1.0 |ComplexBest = 7000000048.0| Time(min) = 0.0771279 GA-PARSIMONY | iter = 21 MeanVal = 0.9150223 | ValBest = 0.972731 | TstBest = 1.0 |ComplexBest = 7000000048.0| Time(min) = 0.0847196 GA-PARSIMONY | iter = 22 MeanVal = 0.9335024 | ValBest = 0.972731 | TstBest = 1.0 |ComplexBest = 7000000048.0| Time(min) = 0.0814945 +------------------------------------+ | GA-PARSIMONY | +------------------------------------+ GA-PARSIMONY settings: Number of Parameters = 2 Number of Features = 13 Population size = 40 Maximum of generations = 50 Number of early-stop gen. = 10 Elitism = 8 Crossover probability = 0.8 Mutation probability = 0.1 Max diff(error) to ReRank = 0.001 Perc. of 1s in first popu.= 0.9 Prob. to be 1 in mutation = 0.1 Search domain = C gamma alcohol malic_acid ash alcalinity_of_ash \ Min_param 0.0001 0.00001 0.0 0.0 0.0 0.0 Max_param 99.9999 0.99999 1.0 1.0 1.0 1.0 magnesium total_phenols flavanoids nonflavanoid_phenols \ Min_param 0.0 0.0 0.0 0.0 Max_param 1.0 1.0 1.0 1.0 proanthocyanins color_intensity hue \ Min_param 0.0 0.0 0.0 Max_param 1.0 1.0 1.0 od280/od315_of_diluted_wines proline Min_param 0.0 0.0 Max_param 1.0 1.0 GA-PARSIMONY results: Iterations = 23 Best validation score = 0.9727309855126027 Solution with the best validation score in the whole GA process = fitnessVal fitnessTst complexity C gamma alcohol malic_acid ash \ 0 0.972731 1 7e+09 51.1573 0.0581044 1 0 1 alcalinity_of_ash magnesium total_phenols flavanoids nonflavanoid_phenols \ 0 1 0 0 1 0 proanthocyanins color_intensity hue od280/od315_of_diluted_wines proline 0 0 0 1 1 1 Results of the best individual at the last generation = Best indiv's validat.cost = 0.9727309855126027 Best indiv's testing cost = 1.0 Best indiv's complexity = 7000000048.0 Elapsed time in minutes = 1.9634766817092892 BEST SOLUTION = fitnessVal fitnessTst complexity C gamma alcohol malic_acid ash \ 0 0.972731 1 7e+09 51.1573 0.0581044 1 0 1 alcalinity_of_ash magnesium total_phenols flavanoids nonflavanoid_phenols \ 0 1 0 0 1 0 proanthocyanins color_intensity hue od280/od315_of_diluted_wines proline 0 0 0 1 1 1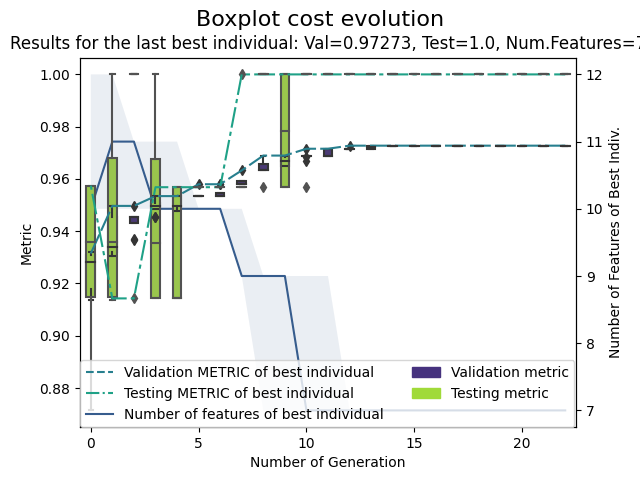
Classification plot
- _crossover(parents, alpha=0.1, perc_to_swap=0.5)
Function for crossover in GAparsimony.
Functions implementing particular crossover genetic operator for GA-PARSIMONY. Method usesfor model parameters Heuristic Blending and random swapping for binary selected features. Modify the attributes: population, fitnessval, fitnesstst and complexity.
- Parameters
parents (list) – A list with two integers that correspond to the indices of the rows of the parents of the current population.
alpha (float, optional) – A tuning parameter for the Heuristic Blending outer bounds [Michalewicz, 1991]. Typical and default value is 0.1.
perc_to_swap (float, optional) – Percentage of features for swapping in the crossovering process. Default value is 0.5.
- _mutation()
Function for mutation in GAparsimony.
Functions implementing mutation genetic operator for GA-PARSIMONY. Method mutes a GAparsimony.pmutation percentage of them. If the value corresponds to a model parameter, algorithm uses uniform random mutation. For binary select features, method sets to one if the random value between [0,1] is loweror equal to GAparsimony.feat_mut_thres. Modify the attributes: population, fitnessval, fitnesstst and complexity.
- _population(type_ini_pop='randomLHS')
Population initialization in GA-PARSIMONY with a combined chromosome of model parameters and selected features. Functions for creating an initial population to be used in the GA-PARSIMONY process.
Generates a random population of GAparsimony.popSize individuals. For each individual a random chromosome is generated with len(GAparsimony.population._params) real values in the range[GAparsimony._min, GAparsimony._max] ` plus `len(GAparsimony.population.colsnames) random binary values for feature selection. random or Latin Hypercube Sampling can be used to create a efficient spread initial population.
- Parameters
type_ini_pop (list, {'randomLHS', 'geneticLHS', 'improvedLHS', 'maximinLHS', 'optimumLHS'}, optional) – How to create the initial population. random optiom initialize a random population between the predefined ranges. Values randomLHS, geneticLHS, improvedLHS, maximinLHS & optimumLHS corresponds with several meth-ods of the Latin Hypercube Sampling (see lhs package for more details).
- Returns
A matrix of dimension GAparsimony.popSize rows and len(GAparsimony.population._params)+len(GAparsimony.population.colsnames) columns.
- Return type
numpy.array
- _rerank()
Function for reranking by complexity in parsimonious model selectionprocess. Promotes models with similar fitness but lower complexity to top positions.
This method corresponds with the second step of parsimonious model selection (PMS) procedure.PMS works in the following way: in each GA generation, best solutions are first sorted by their cost,J. Then, in a second step, individuals with less complexity are moved to the top positions when theabsolute difference of their J is lower than aobject@rerank_errorthreshold value. Therefore, theselection of less complex solutions among those with similar accuracy promotes the evolution ofrobust solutions with better generalization capabilities.
- Returns
A vector with the new position of the individuals
- Return type
numpy.array
- _selection(*args, **kwargs)
Function for selection in GAparsimony.
Functions implementing selection genetic operator in GA-PARSIMONY after parsimony_rerankprocess. Linear-rank or Nonlinear-rank selection (Michalewicz (1996)). The type of selection is specified with the model selection attribute, it can be: linear, nlinear or random. Modify the attributes: population, fitnessval, fitnesstst and complexity.
- fit(X, y, iter_ini=0)
A GA-based optimization method for searching accurate parsimonious models by combining feature selection, model tuning, and parsimonious model selection (PMS). PMS procedure is basedon separate cost and complexity evaluations. The best individuals are initially sorted by an errorfitness function, and afterwards, models with similar costs are rearranged according to their modelcomplexity so as to foster models of lesser complexity.
- Parameters
X (pandas.DataFrame or numpy.array) – Training vector.
y (pandas.DataFrame or numpy.array) – Target vector relative to X.
iter_ini (int, optional) – Iteration/generation of GAparsimony.history to be used when model is pretrained. If iter_ini==None uses the last iteration of the model.
- importance()
Percentage of appearance of each feature in elitist population.
Shows the percentage of appearance of each feature in the whole GA-PARSIMONY process butonly for the elitist-population. If it is assigned, it returns a dict if not displayed on the screen.
- Returns
A numpy.array with information about feature importance.
- Return type
numpy.array
- plot(min_iter=None, max_iter=None, main_label='Boxplot cost evolution', steps=5, size_plot=None, *args)
Plot of GA evolution of elitists.
Plot method shows the evolution of validation and testing errors, and the number of model features selected of elitists. White and grey box-plots represent validation and testing errors of elitists evo-lution, respectively. Continuous and dashed-dotted lines show the validation and testing error ofthe best individual for each generation, respectively. Finally, the shaded area delimits the maximumand minimum number of features, and the dashed line, the number of features of the best individual.
- Parameters
min_iter (int, optional) – Min GA iteration to visualize. Default None.
max_iter (int, optional) – Max GA iteration to visualize.Default None.
main_label (str, optional) – Main plot title.Default ‘Boxplot cost evolution’.
steps (int, optional) – Number of divisions in y-axis. Default 5.
size_plot (tuple, optional) – The size of the plot. Default None
- predict(X)
Predict result for samples in X.
- Parameters
X (numpy.array or pandas.DataFrame) – Samples.
- Returns
A numpy.array with predictions.
- Return type
numpy.array
- summary(**kwargs)
Summary for GA-PARSIMONY.
Summary method for class GAparsimony. If it is assigned, it returns a dict if not displayed on the screen.
- Returns
A dict with information about the GAparsimony object.
- Return type
dict